News Story
Monje-Galvan Presents Work on Molecular Dynamics Simulations of Protein Binding
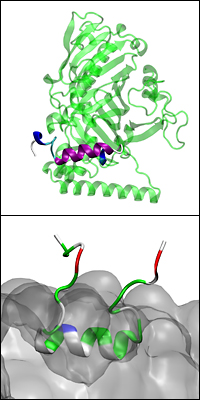
Above: A model of the full Osh4 protein with the ALPS-like motif highlighted in purple and blue. Below: The ALPS-like motif embedded on a charged membrane (bulk shown in grey) and interacting with its lipid headgroups.
University of Maryland graduate student Viviana Monje-Galvan (Department of Chemical and Biomolecular Engineering [ChBE]) received the Chemical Society of Washington’s travel award to support her trip to the 249th National Meeting of the American Chemical Society (ACS). Monje-Galvan, advised by ChBE associate professor Jeffery Klauda, headed to Denver, Co. in late March to present her molecular dynamics simulations of model cell membranes and peptide interactions as the first step in a study of a lipid transport protein in yeast.
Cell membranes, she explains, need to contain and maintain the correct concentration or amount of lipids (fats) to keep the cell healthy and alive. She is using advanced computer simulations to study the behavior of a protein called Osh4, which is known to transfer specific lipids between organelles in yeast cells. Although Osh4 has been the subject of experimental studies, little is known about how it does its job. Monje-Galvan’s research focuses on discovering what forces drive and stabilize the protein’s process of attaching itself to cell membranes.
“Although Osh4 is a protein in yeast, it has similarities to a protein in humans that's involved in cholesterol transport and has been linked to the activity of cancer cells,” says Klauda. "But that human prontein still not well understood. Hopefully our work using Osh4 as a homologue will give us some insight.” Understanding its role in yeast, he adds, could also provide beneficial information to research groups that are developing cancer therapies.
In this initial study, Monje-Galvan and Klauda focused on only a portion of the Osh4 protein, a curvature-sensing peptide called the ALPS-like motif. As it nears a cell, the ALPS-like motif scans the surface of its membrane, looking for areas in which the lipids that form it are not tightly packed together, a phenomenon known as a packing defect. These defects are more likely to occur where the membrane is curved and the hydrophobic lipid tails are more exposed to the peptide. The ALPS-like motif is drawn to these areas because the extra space between the lipid heads allows it to work its way into the membrane’s inner core. Monje-Galvan and Klauda were able to study this behavior using all-atom simulations of the peptide with simple and complex model membranes.
“In the case of electrostatically neutral model membranes, the peptide interacted poorly with the cell membrane due to weak electrostatic contacts and too-short simulations, compared to the actual binding time scale of one microsecond,” says Monje-Galvan. But, she adds, the peptide inserted itself into the membrane’s hydrophobic core (the area where the lipid tails reside) when they ran longer simulations with settings that incorporated surface tension and a charged membrane. “We introduced surface tension into our simulations, basically stretching the membranes, to produce more packing defects.”
The simulations revealed that the ALPS-like motif inserts horizontally into the inside of the membrane only when the lipid packing defect is large enough for the hydrophobic amino acids in its structure to ‘see’ the hydrophobic core of the membrane. The pair also discovered that the peptide amino acids with ring structures help stabilize its insertion and anchor it to the membrane’s core.
“Our results allow us to move forward to study the interaction of the entire Osh4 protein with our complex model membranes,” says Monje-Galvan. “The final goal is to understand Osh4’s binding mechanism to a membrane and hopefully also how it takes up and transports lipids to another membrane.”
Published March 30, 2015









